https://github.com/savastakan/dnmso#Gap
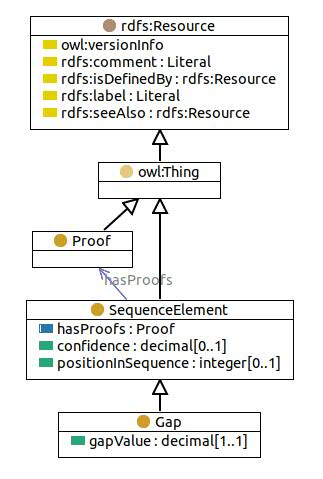
Class 'Gap; an m/z range that couldn't be assigned an AminoAcid or a ModAminoAcid. '
type
-
 |
Class [owl:Class] |
|
comment
-
 |
For de novo sequencing results, it often occurs that some parts of the sequence cannot be supported with much evidence. Instead of storing an arbitrary subsequence, a gap of appropriate m/z range (monoisotopic mass) can be inserted in the Sequence. The gapValue represents the size of the gap and is obligatory.
|
|
label
-
 |
Gap; an m/z range that couldn't be assigned an AminoAcid or a ModAminoAcid.
|
|
subClassOf
-
-
|
Generated with TopBraid Composer
by TopQuadrant, Inc.

